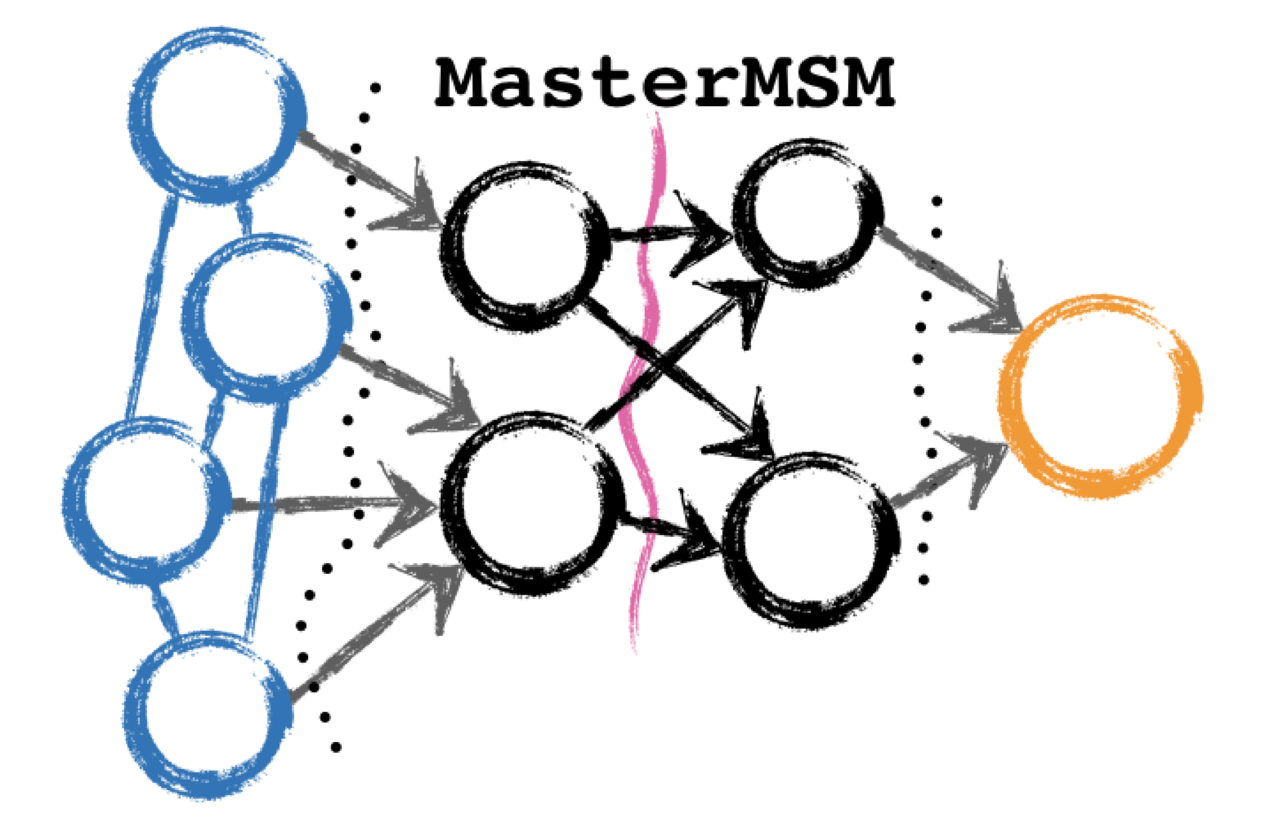
Welcome to MasterMSM’s documentation!¶
MasterMSM is a Python package for generating Markov state models (MSMs) from molecular dynamics trajectories. We use a formulation based on the chemical master equation. This package will allow you to:
- Create Markov state / master equation models from biomolecular simulations.
- Discretize trajectory data using dihedral angle based methods useful for small peptides.
- Calculate rate matrices using a variety of methods.
- Obtain committors and reactive fluxes.
- Carry out sensitivity analysis of networks.
We have written a paper on MasterMSM that briefly describes some of the code capabilities. The MasterMSM code is hosted in Github. Active development of the MasterMSM code takes place using the git version control system.
Contents: